Siemens Prisma 3T walkthrough
Organization
The purpose of the ReproIn effort is to automate as much as possible, requiring just minimal amount of information entry at the scanner sufficient to have collected session be placed correctly within a hierarchy of the datasets, get new data converted to BIDS, and (optionally) be placed under the version control by DataLad.
To achieve that first we should make it possible to make DICOMs cary information about the Investigator (e.g. a PI of the study or a mentor), possibly corresponding Experimenter (student/assistant or Investigator himself), and the study (in our case ID-name) itself.
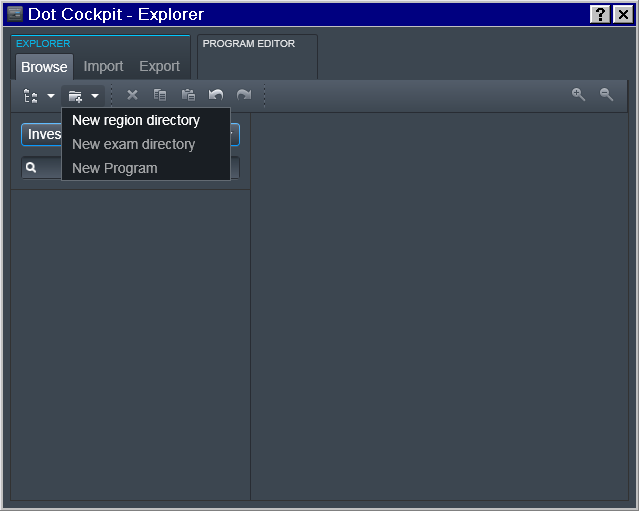
Tree -> Investigator
To accomplish that, in the Dot Cockpit we first created a dedicated tree for each Investigator:
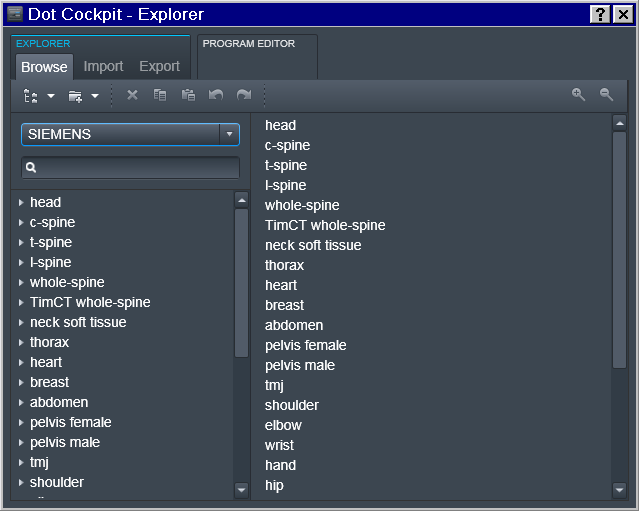
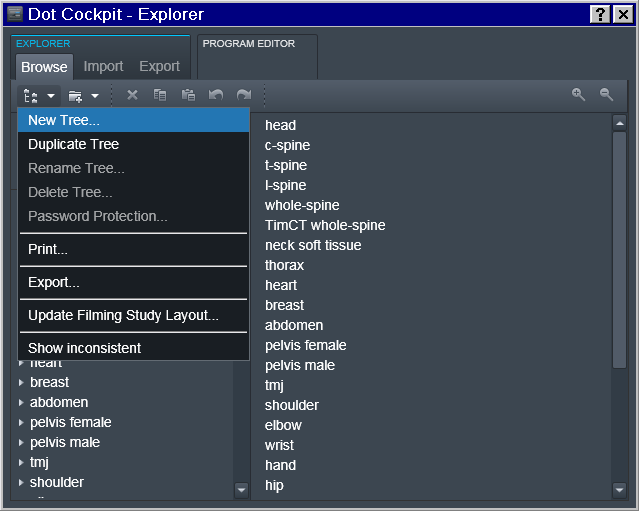
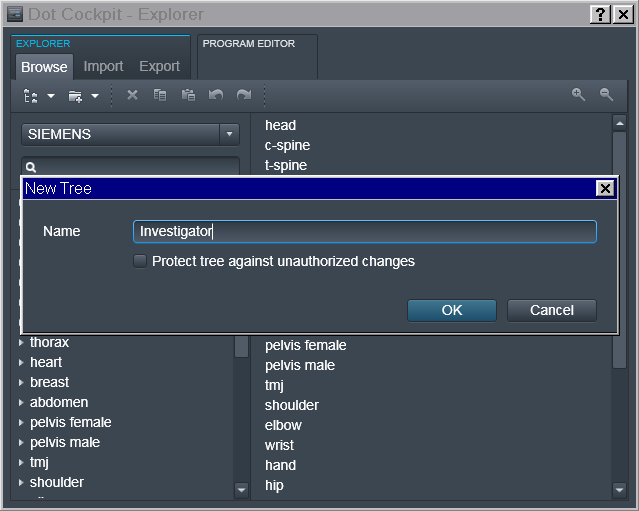
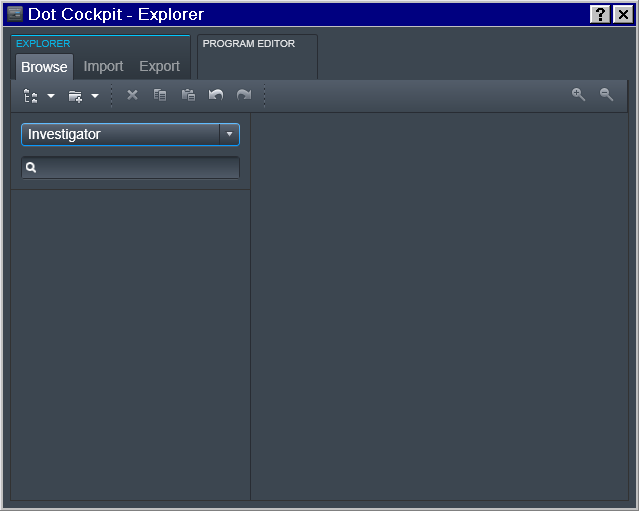
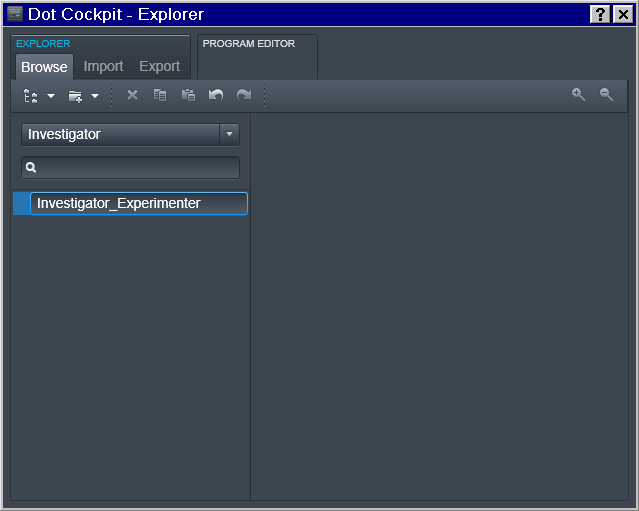
Region -> Experimenter
Then for a specific Experimenter who is responsible for the study we
defined the 2nd level (Region) entry as a join Investigator_Experimenter
entry
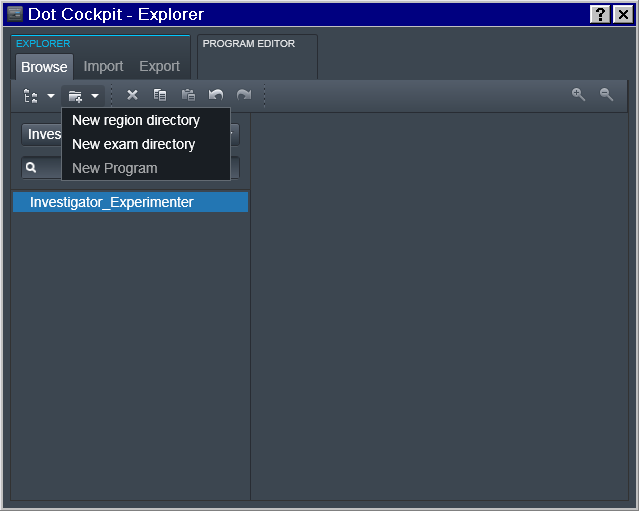
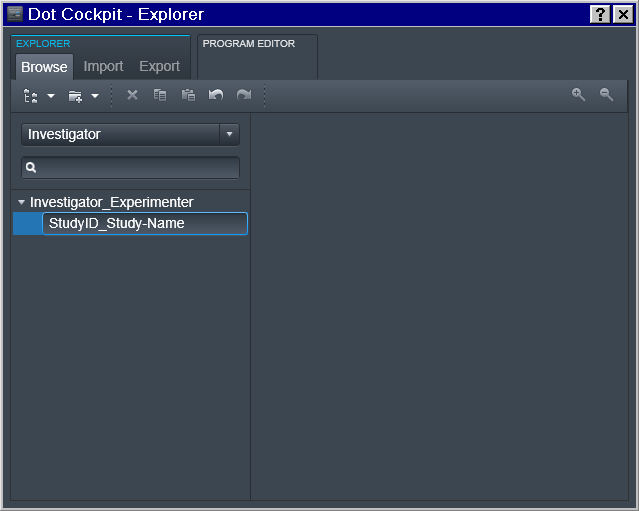
Exam -> Study
And then at the 3rd level of the Exam we define the study.
Note: you could have multiple entries at any of those levels (e.g. multiple Experimenters working for the same PI; or multiple studies for the same Experimenter):
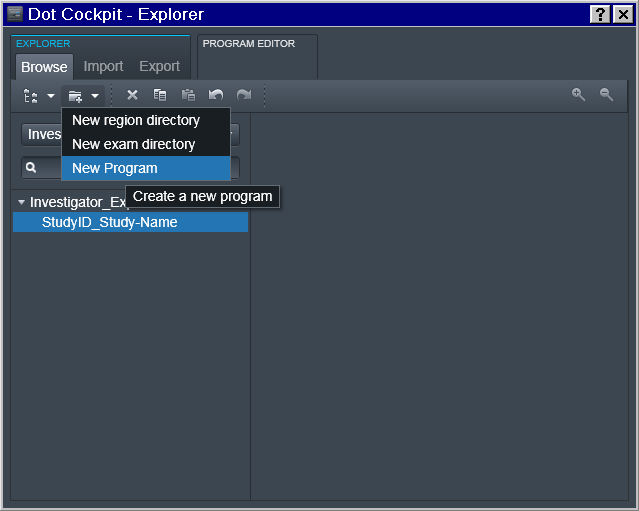
New Program
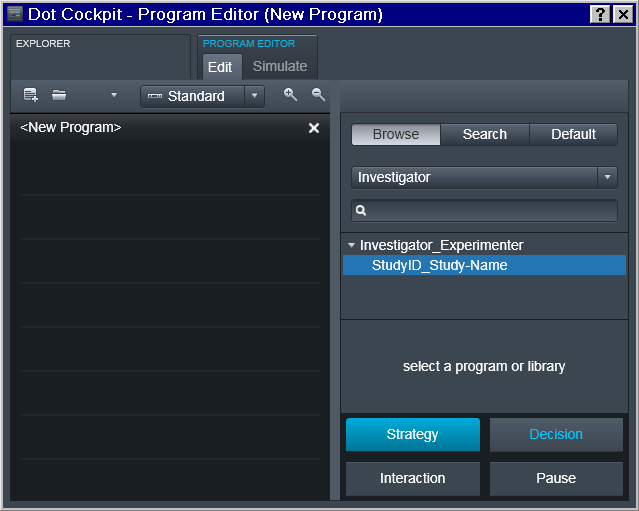
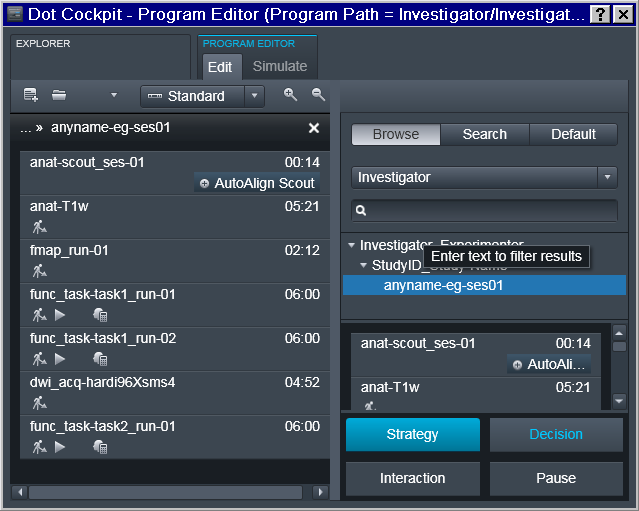
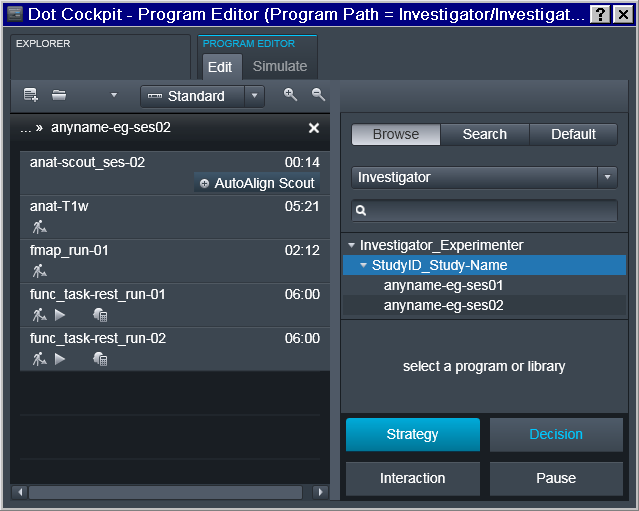
Now it is possible to finally define Program(s) with the desired sequence of protocols:
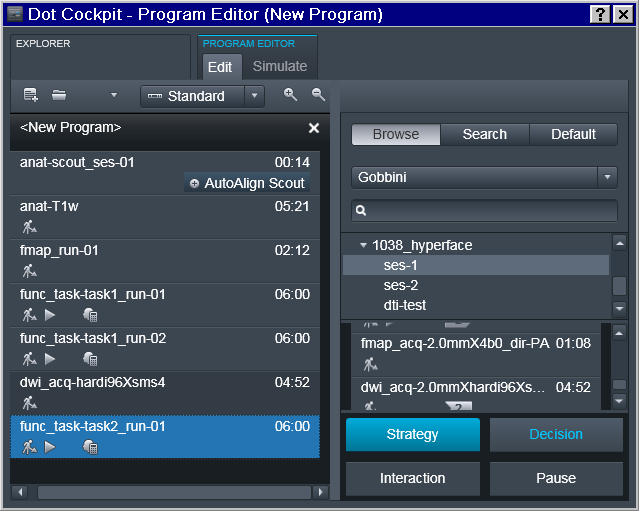
Note that the sequence names follow reproin specification. So in the example below it is intended for the first session of a study collecting a T1 anatomical, a fieldmap, two runs for task1 functional sequence, then a diffusion image, and completes with a single run for the functional task2. Those sequences could be copied from prior/other studies which might have already followed the naming convention and otherwise have desired settings:
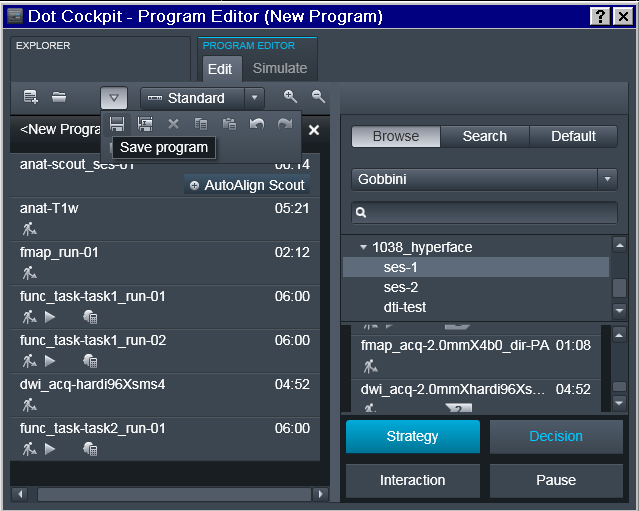
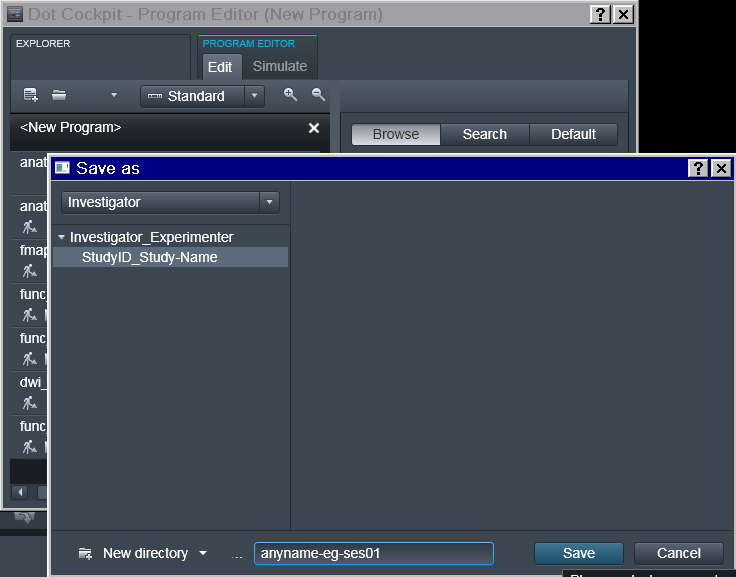
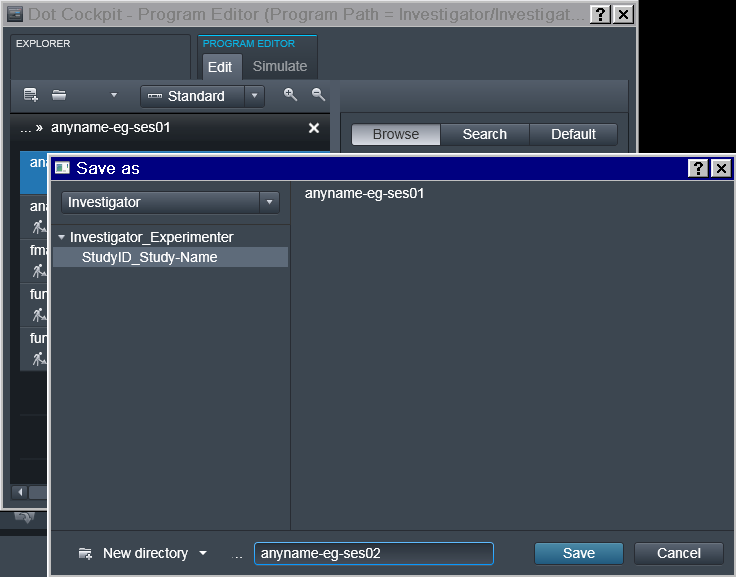
Program should be saved under some descriptive (but otherwise arbitrary)
name. Note that if the study requires multiple scanning sessions, it is
useful to use _ses- suffix to right away depict for which session a
particular program is intended.
Next Session
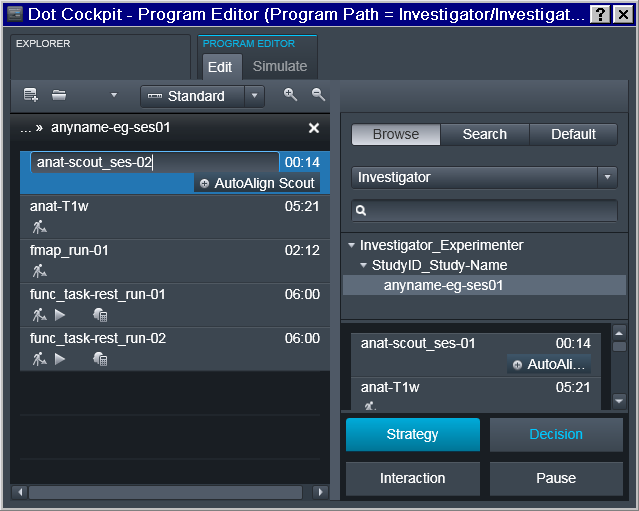
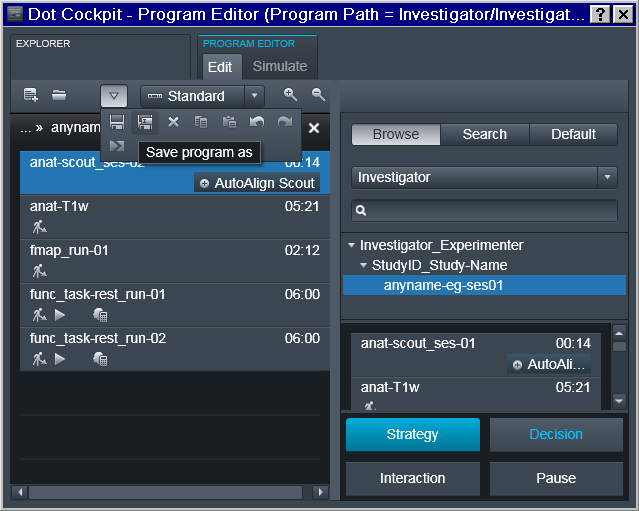
Next session program should acquire session marker within the name of the scout sequence and be saved separately:
New Accession
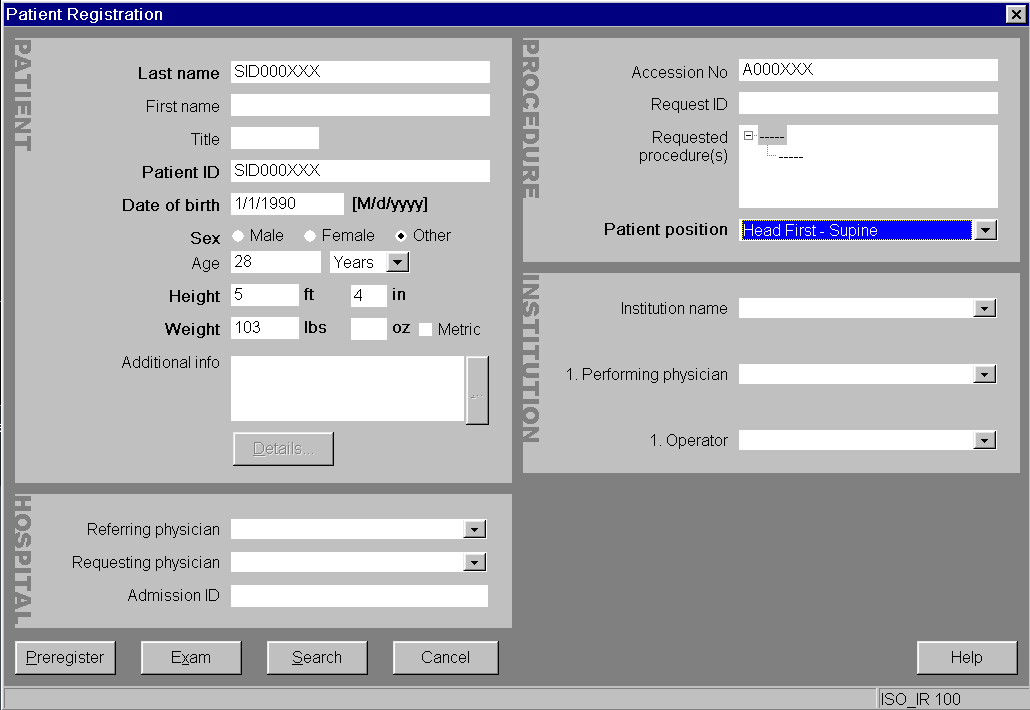
Register a (New) Subject
In the exam card we enter no personal information to avoid leakage through
DICOMs. All personal data for a given subject id is stored elsewhere
(Age and Sex from the exam card though are used to autofill up
age and sex fields within participants.tsv):
Choose the Investigator in the Tree
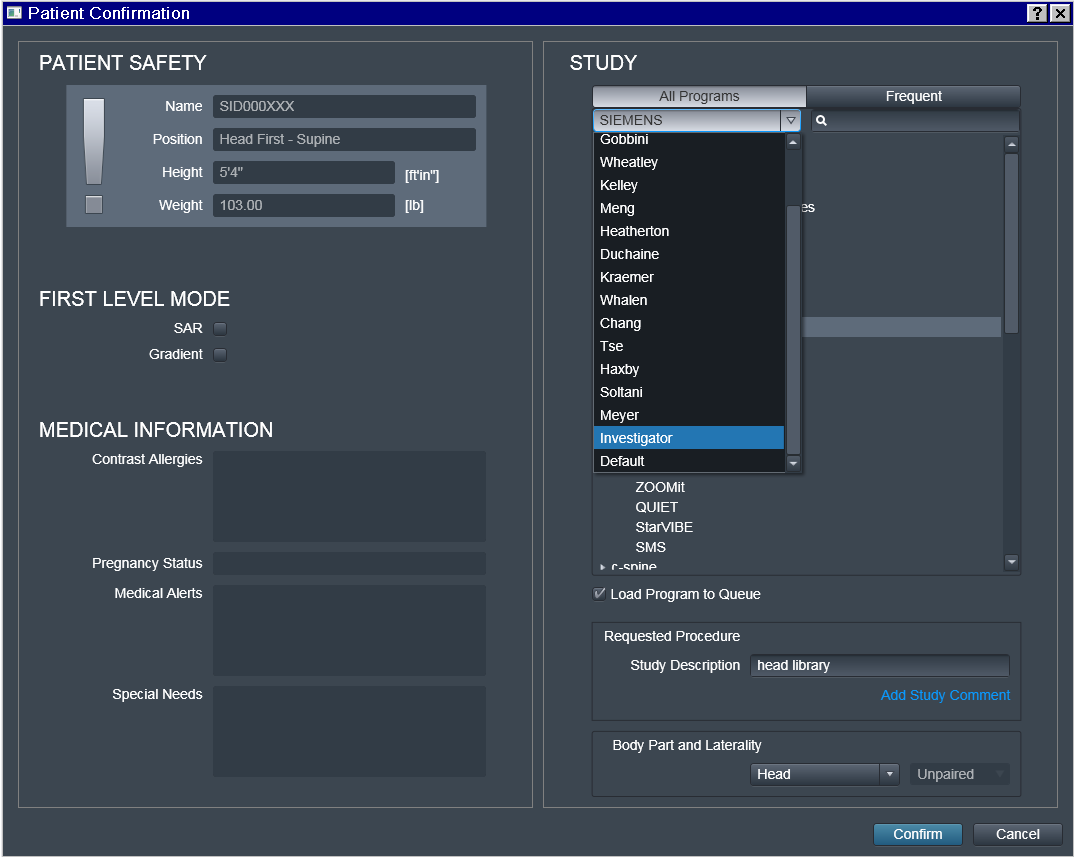
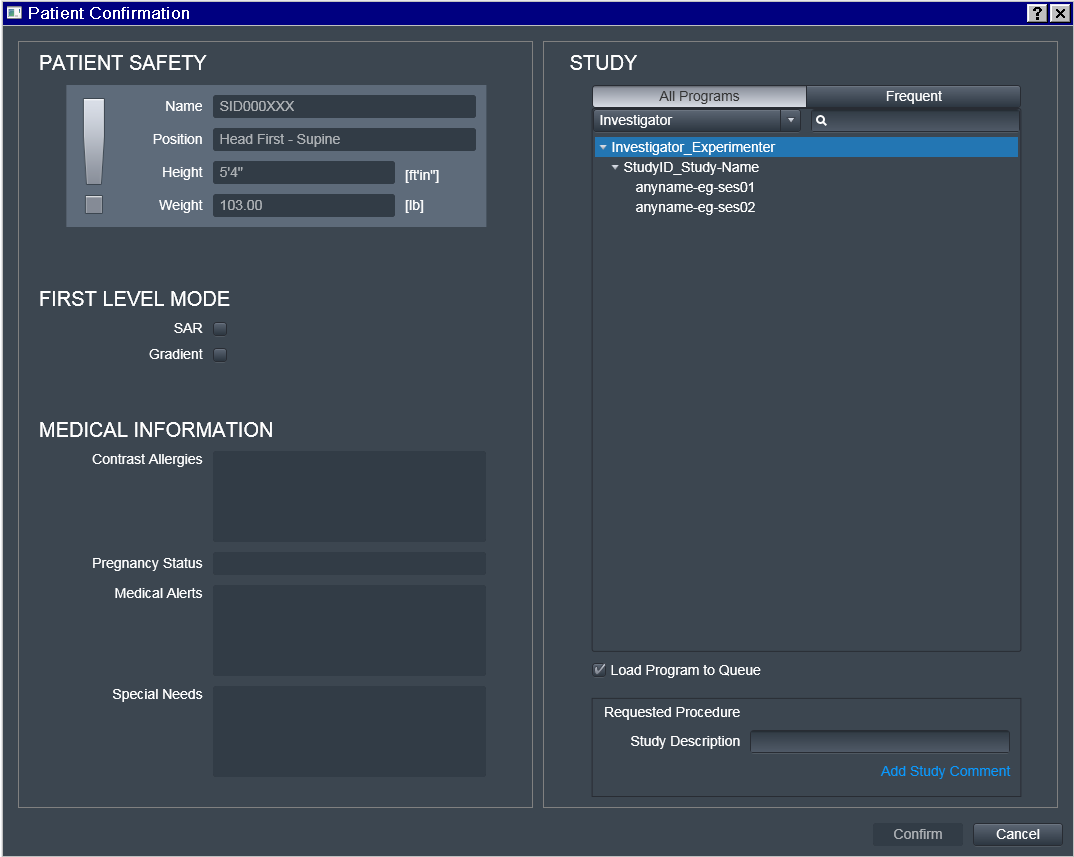
Choose the desired Program
That is where some "magic" (i.e. automation) happens:
Investigator_Experimenter and StudyID_Study-Name fields get copied
by the UI into the Study Description field which later is included
within transmitted DICOM headers.
Note also that only the Investigator_Experimenter and
StudyID_Study-Name fields get copied into the Study Description. The
Program name (such as anuname-eg-ses01) is not copied, and
appears nowhere within DICOM precluding its utility for automation
(hence anyname was chosen here for this example).
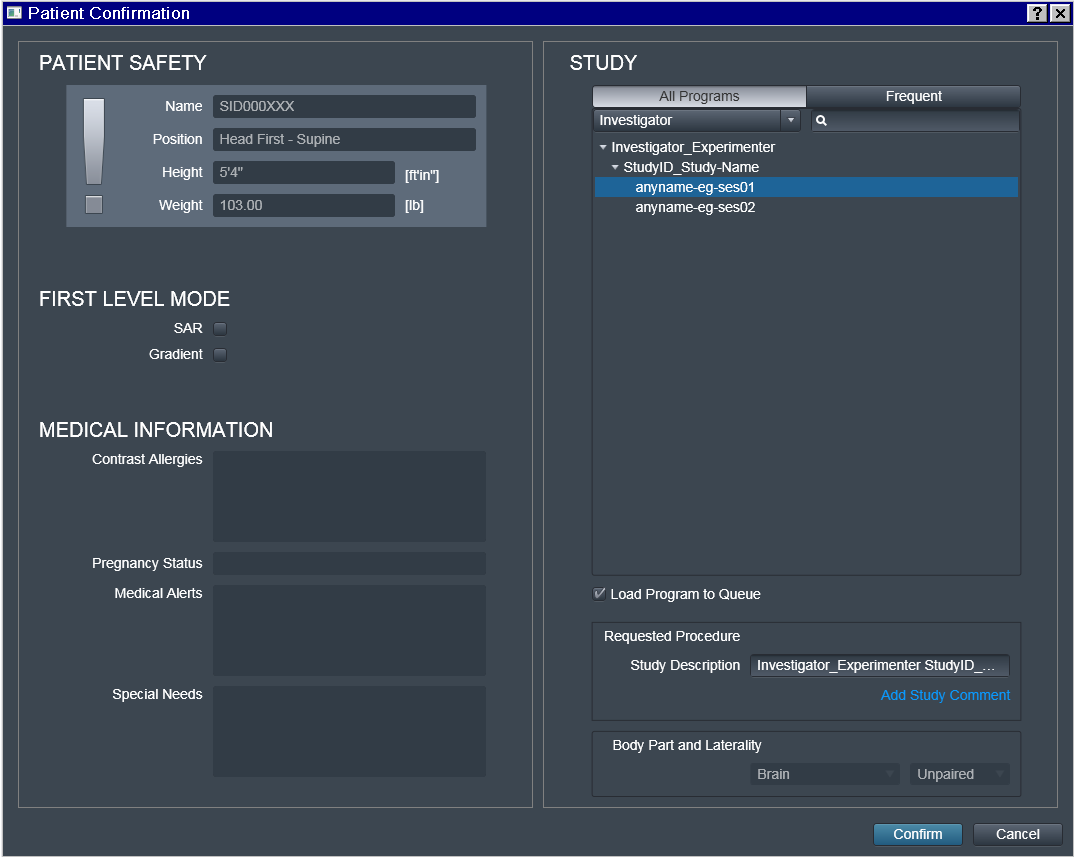
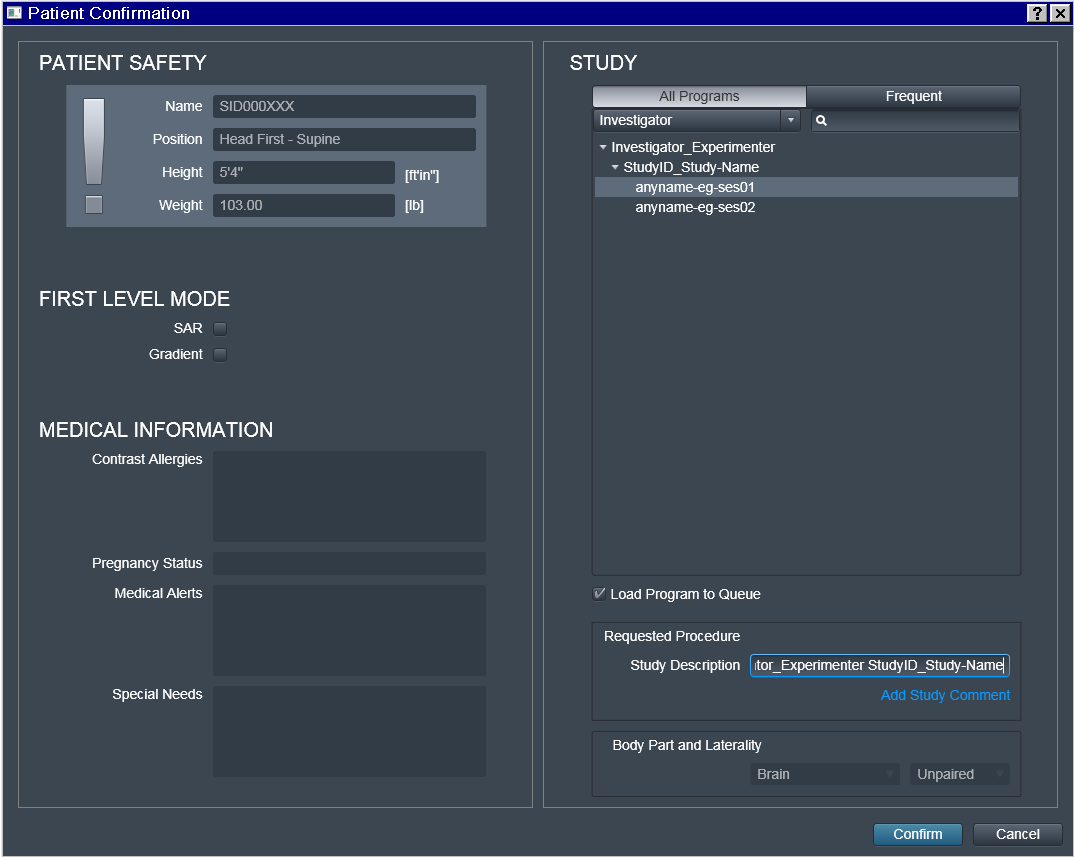
Do not edit Study Description, unless you really need to and can
guarantee consistency. This location will determine the location of
the dataset on the file system within the hierarchy of (DataLad if ran
with --datalad option) datasets. Having it this fully automated
guarantees that for the next subject/session, data will be placed into
the same dataset without the need to specify target location manually,
and thus preventing possible human errors.
Interrupted Scan
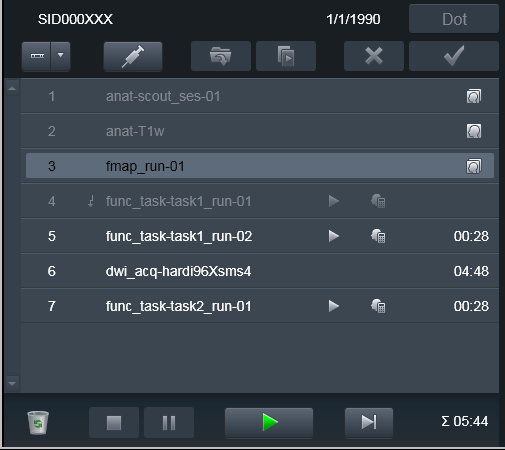
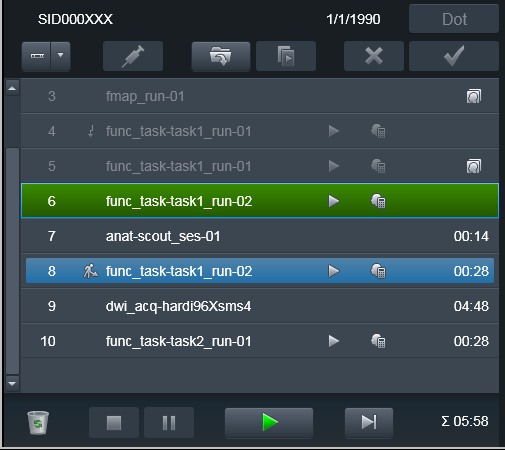
As you can see in the following example, we have interrupted
func_task-task1_run-01 functional scan, may be because our phantom
fell asleep. Our scanner console is configured to transmit data
immediately upon succesful volume was collected, so those volumes were
already transmitted to PACS:
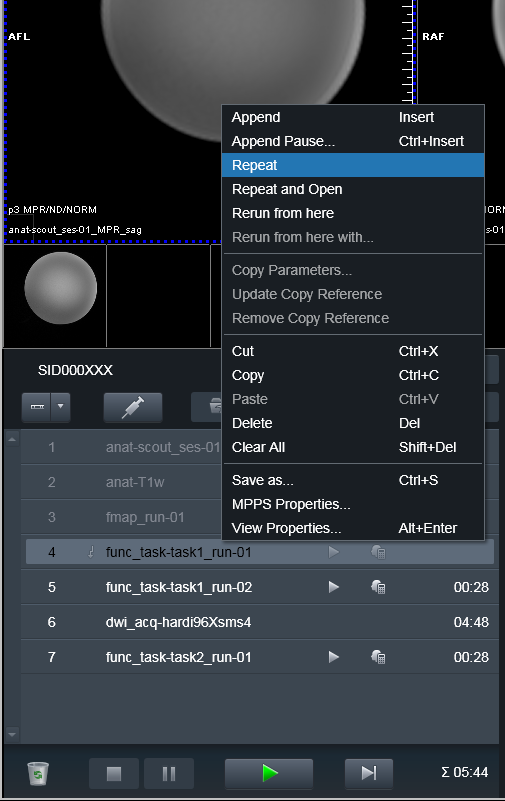
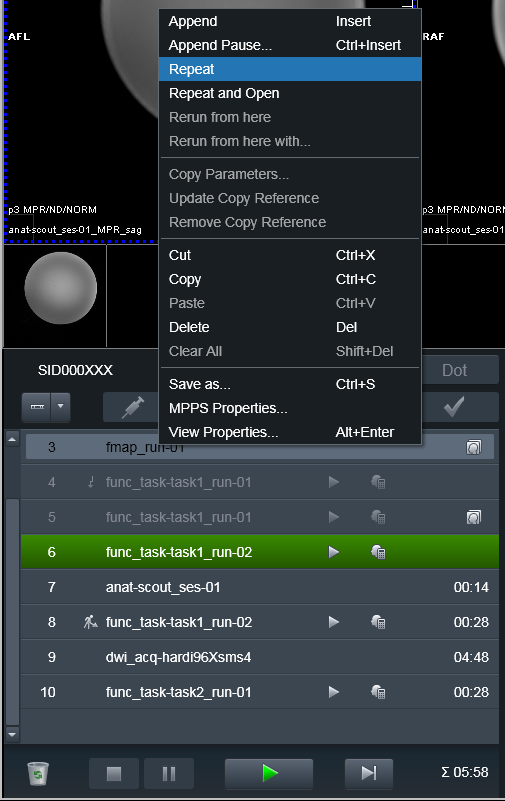
To make heudiconv reproin heuristic figure out that the run was
canceled or otherwise needs to be discarded, just Repeat the run
without changing anyting in its name!:
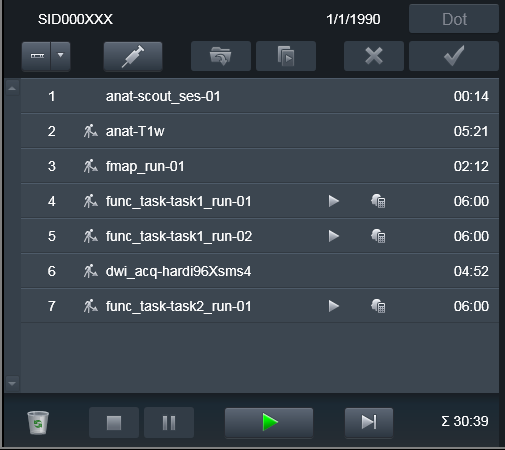
Then, upon conversion, the earlier (e.g. canceled) scans will also be
converted, but assigned __dupX suffix (X is an incrementing integer to
allow for possibly multiple canceled runs). This way data curator could
inspect those volumes and if everything matches the notes/sanity checks,
remove those __dupX files. Meanwhile you can proceed with completing
your Program of scans:
Interrupted Program
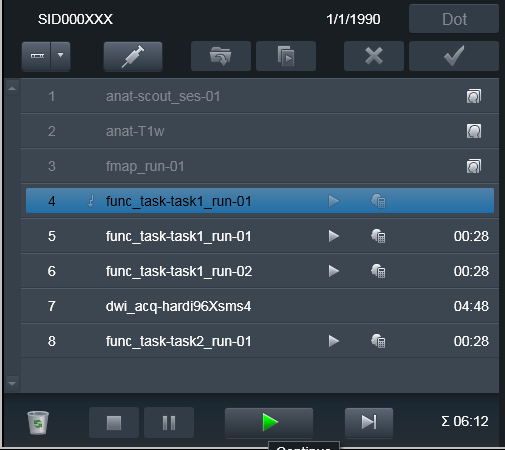
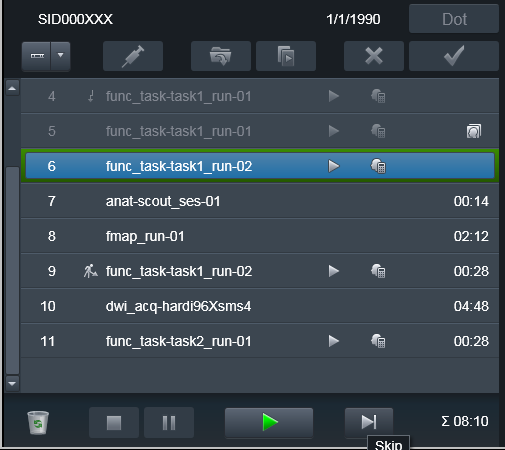
Some times it is necessary to take subject outside of the scanner, and
bring him back later to finish the scanning session. Typically some volumes
(e.g. at least scouts and fieldmaps) need to be reran. Because it would be
desired to keep both version of the files -- from both original scanning
session and the continued one -- you would need to Repeat those scans
but this time assign them a new suffix. E.g., you could assign the _run-
suffix matching the run_ suffix of the next functional run, so it would
make it easier later on to associate fieldmaps with the corresponding
functional run file(s).
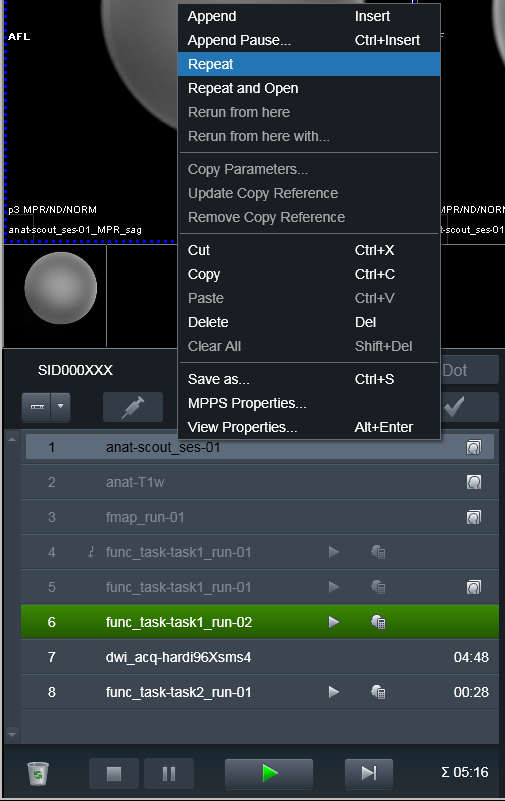
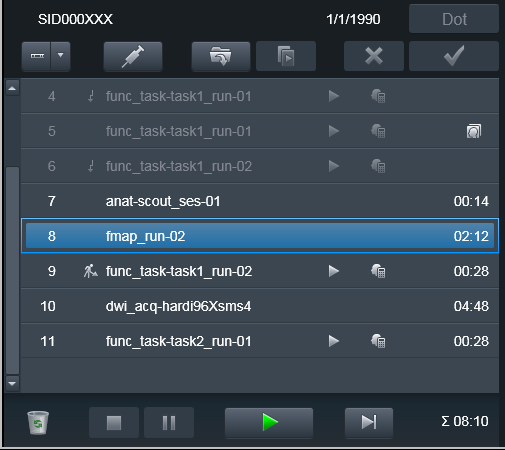
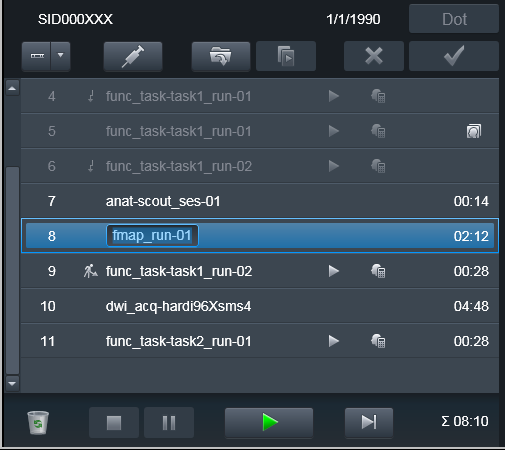
Done again:

Data was transmitted to PACS and ready for processing using heudiconv
-f reproin.