DataLad
The DataLad Handbook at http://handbook.datalad.org is the ultimate introduction into DataLad.
An example of sample analysis using DataLad & containers
This is an example on how to organize your study as DataLad dataset and use datalad-container extension to facilitate efficient and reproducible research:
# Create a dataset where we will keep all bits and pieces of a study
# With -c text2git we instruct to have data files to go to annex, while
# text files (code, docs, scripts) to git
datalad create -c text2git 1021_actions
cd 1021_actions
# Install a (sub)dataset with (Singularity) containers
datalad install -d . http://github.com/repronim/containers
# Install ReproIn'ed dataset from rolando
datalad install -d . -s rolando.cns.dartmouth.edu:/inbox/BIDS/Haxby/Sam/1021_actions bids
# Getting only a sample data (1 subject) for demonstration here
datalad get bids/sub-sid000005
Meanwhile, have a look at
- https://github.com/ReproNim/containers/#a-typical-workflow
- a bit too convoluted ATM example with either
datalad runorreproman runfor scheduling parallel execution across cluster: https://github.com/ReproNim/reproman/pull/438 - and possible future answers on https://neurostars.org/t/using-fmriprep-with-datalad-containers-run/5327
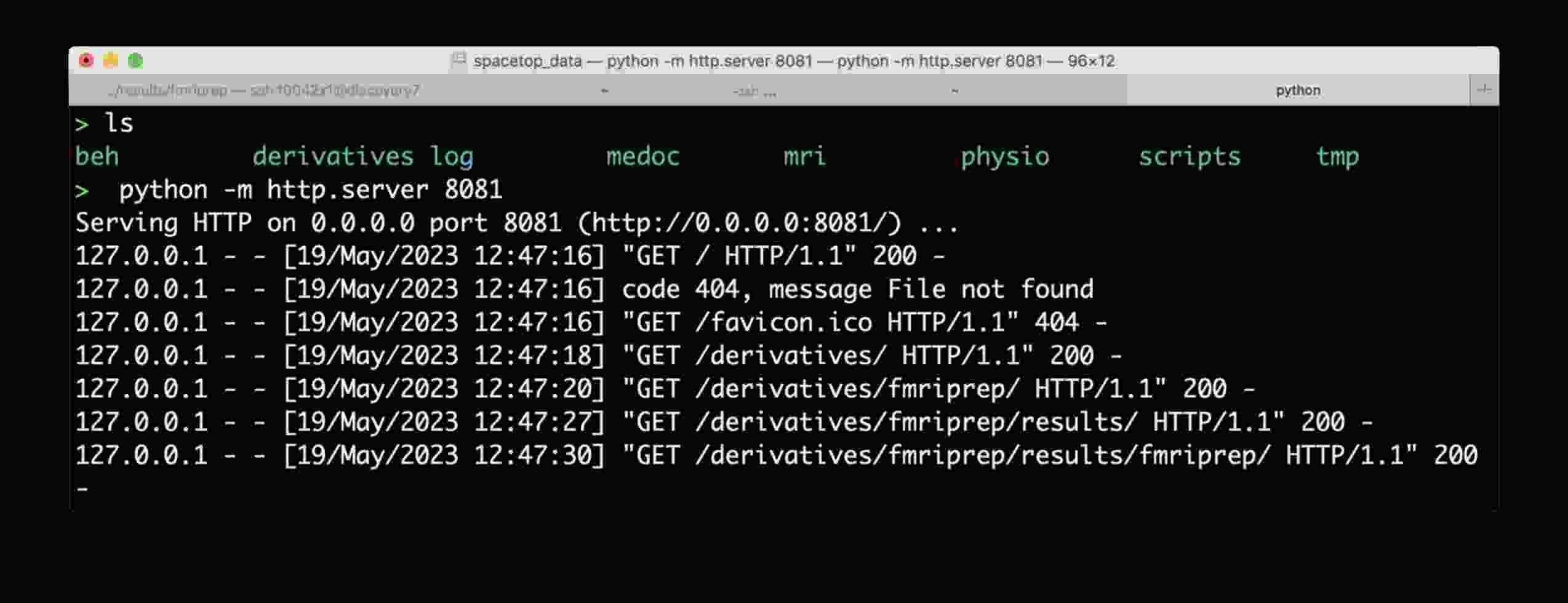
How to view mriqc/fmriprep/etc DataLad'ified results in a browser
The problem: web browser de-references symlinks, which leads .html into a .git/annex/objects subfolder and thus makes it impossible to see the images.
To overcome this, start a simple web server, e.g. as provided by Python itself, and navigate to the file of interest:
- Run this line of code on local directory (e.g., where you have mounted discovery directory, or have a local clone with data fetched):
python -m http.server 8081 - Copy the URL it prints out (typically http://0.0.0.0:8081/) into your web browser address bar
- Now you should be able to access HTML outputs with embedded images at that address -- just navigate to .html file of interest and open it.
DataLad way
Make sure you have git configured, in that it knows about you.
If you have output to running git config user.name command, then most likely you are all set.
If it comes out empty, please do
git config --global user.name "First Last"
git config --global user.email "someemail@address"
while replacing those values with your name and email.
If you are doing it to discovery HPC, please first checkout the next section of this documentation below (Discovery filesystem) on how to configure your git for Discovery's ACL filesystem, unless you know that it is pure POSIX.
Then it is recommended to create a directory for your study first, e.g. mkdir ID_name where ID_name are the same as on rolando, e.g. 0001_dbic-animals, and cd into it, e.g.:
mkdir 0001_dbic-animals
cd 0001_dbic-animals
Use datalad install command
to obtain dataset, and then datalad get to
obtain specific files. If you are greedy, add -r to get full hierarchy of datasets, and/or -g
to immediately also fetch all data files
datalad install -s your-login-on-rolando@rolando.cns.dartmouth.edu:/inbox/BIDS/dbic/0001_dbic-animals dbic
cd dbic
datalad get -J4 sub-* # to get only converted data, without tarballs etc
Later upgrades to fetch new data (subjects etc) could be done via
datalad update --how merge -r